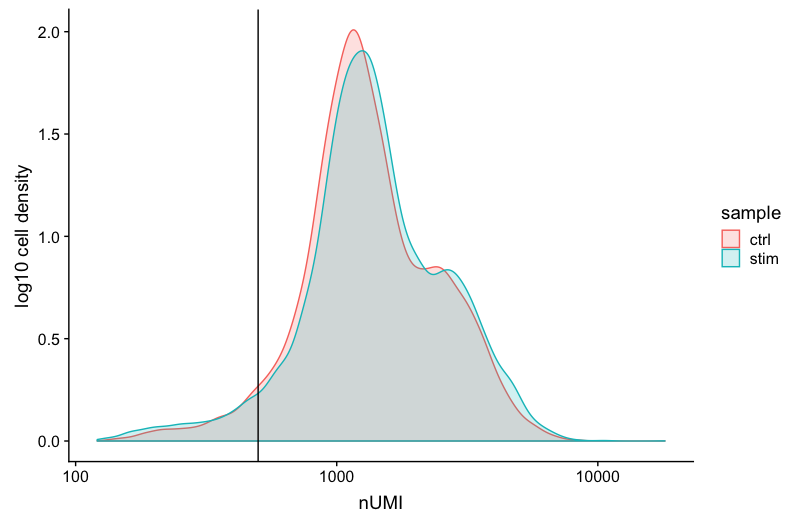
Umi Scrna Seq . unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise.
from hbctraining.github.io
unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise.
Singlecell RNAseq Quality Control Analysis Introduction to single
Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise.
From www.rna-seqblog.com
Comparative analysis of singlecell RNAsequencing methods RNASeq Blog Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From www.researchgate.net
The effect of sequencing depth and cell number on single cell UMI Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From dmgatti.github.io
Quality Control of scRNASeq Data Single Cell RNAseq Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From www.frontiersin.org
Frontiers A machine learning framework for scRNAseq UMI threshold Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From hbctraining.github.io
Generation of count matrix Introduction to singlecell RNAseq Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From labs.wsu.edu
Overview Analyzing scRNAseq Winuthayanon Lab Washington State Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From www.genomescan.nl
Increasing sequencing accuracy with unique molecular identifiers Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From dnatech.genomecenter.ucdavis.edu
What are UMIs and why are they used in highthroughput sequencing Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From www.researchgate.net
UMI coding for individual plasma DNA molecules to enhance the accuracy Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From www.rna-seqblog.com
The evolution of singlecell RNA sequencing technology and application Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From www.researchgate.net
ScRNAseq profiling of the in vitro PGCLC differentiation system a Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From www.researchgate.net
UMI incorporation into RNAseq. a Overall workflow. Schematic of a read Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From hbctraining.github.io
Singlecell RNAseq Quality Control Analysis Introduction to single Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From ucdavis-bioinformatics-training.github.io
Flowchart of the scRNAseq analysis Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From frontlinegenomics.com
An overview of singlecell RNA sequencing and spatial transcriptomics Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From www.rna-seqblog.com
A cost effective 5′ selective single cell transcriptome profiling Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From gtpb.github.io
9 Understand specificies of differential gene expression in single Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.
From www.researchgate.net
Single cell RNAseq study design and initial cell grouping. (A) Scheme Umi Scrna Seq unique molecular identifiers (umis) remove duplicates in read counts resulting from polymerase chain reaction, a major source of noise. Umi Scrna Seq.